Reference: Nature, 2012,vol 481, p.39
Title: A sensing array of radically coupled genetic 'biopixels'
Authors: A. Prindle, P. Samayoa, I. Razinkov, T. Danino, L. S. Tsimring, J. Hasty
Review by: Anna Venancio-Marques
Title: A sensing array of radically coupled genetic 'biopixels'
Authors: A. Prindle, P. Samayoa, I. Razinkov, T. Danino, L. S. Tsimring, J. Hasty
Review by: Anna Venancio-Marques
The authors show how an array of bacterias can be used as sensor for arsenite.
1. Choice of sensing array: synchronizing millions of bacteria
The idea is to have a biosensor, for instance a bacteria, that is will report a change in its environment by modifying its behavior, in particular its production of the proteins. However, instead of relying on the response of an individual bacteria to a change in its environment, the authors synchronize the response of several million bacteria in order to have an averaged and more reliable signal.
The bacteria are confined in chambers and separated by PDMS walls. They communicate thanks to the production of H2O22O2, a molecule that activates a specific promoter, the bacteria express a protein, GFP, that will, as it is irradiated, lead to the production of new H2O2. The bacteria can therefore become synchronized over long distances (2.4 cm) and a fluorescent read-out show the bacteria in presence of H2O2.
1. Choice of sensing array: synchronizing millions of bacteria
The idea is to have a biosensor, for instance a bacteria, that is will report a change in its environment by modifying its behavior, in particular its production of the proteins. However, instead of relying on the response of an individual bacteria to a change in its environment, the authors synchronize the response of several million bacteria in order to have an averaged and more reliable signal.
The bacteria are confined in chambers and separated by PDMS walls. They communicate thanks to the production of H2O22O2, a molecule that activates a specific promoter, the bacteria express a protein, GFP, that will, as it is irradiated, lead to the production of new H2O2. The bacteria can therefore become synchronized over long distances (2.4 cm) and a fluorescent read-out show the bacteria in presence of H2O2.
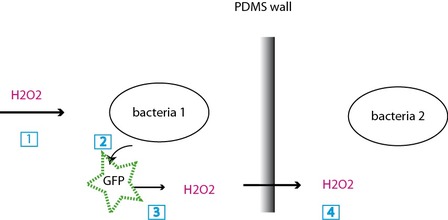
(1) H2O2 signal, (2)the bacteria 1 starts expressing GFP, (3) upon exposure, the GFP generates H2O2, (4) H2O2 crosses the PDMS wall and starts the same chain for bacteria 2.
2. Two detection methods
The authors use the synchronized bacteries as sensors for arsenite.
In the first case (fig 2a), a modification in frequency and amplitude of the oscillations can be linked to a change in arsenite concentration (from 0.25 to 1 uM). Here the expression of the GFP is enhanced in the presence of arsenite. In the absence of arsenite, there is an oscillating fluorescent signal. Its frequency and amplitude is modified in the presence of arsenite.
In the second case (fig 2b) an ON/OFF system with a tunable detection limit is achieved. The circuit is modified, and arsenite is responsible for the synthesis of a molecule that in turns the promoter leading to GFP expresison. In the absence of arsenite, there is no signal, whereas in its presence the oscillating behaviour is recovered.
The authors use the synchronized bacteries as sensors for arsenite.
In the first case (fig 2a), a modification in frequency and amplitude of the oscillations can be linked to a change in arsenite concentration (from 0.25 to 1 uM). Here the expression of the GFP is enhanced in the presence of arsenite. In the absence of arsenite, there is an oscillating fluorescent signal. Its frequency and amplitude is modified in the presence of arsenite.
In the second case (fig 2b) an ON/OFF system with a tunable detection limit is achieved. The circuit is modified, and arsenite is responsible for the synthesis of a molecule that in turns the promoter leading to GFP expresison. In the absence of arsenite, there is no signal, whereas in its presence the oscillating behaviour is recovered.
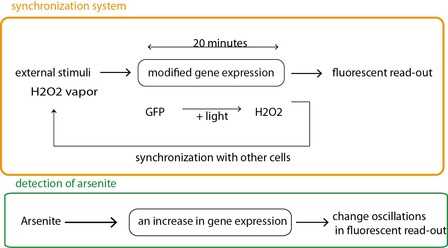
Fig 2a. The synchronizing circuit (in orange) is slightly modified to respond to an arsenite concentration of 0.25 to 1 uM (scheme in green). Arsenite leads to the activation of a supplementary promoter allowing for the expression of GFP.
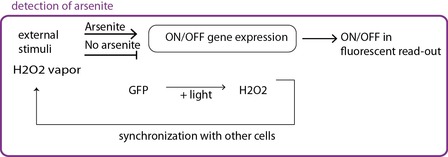
Fig 2b. The circuit itself is modified, the promoter that lead to GFP expression can only be activated in the presence of arsenite.
3. Future challenges
The synchronized bacteries provide a reliable sensor with reduced noise signal and an smooth average response to external stimuli.
However the usefulness of biological systems in electronic-like networks faces many challenges:
- In real-world applications, the big microscopy equipment will have to be scaled down.
- The time lapse between the stimuli and the change in read-out from the system must be reduced. Here the minimum response time is limited by the expression of a protein, which takes about 20 minutes.
- The nature of the read-out needs to be optimized: obtaining for instance an electronic rather than an optical signal, with bacteries emitting electrons directly.
The synchronized bacteries provide a reliable sensor with reduced noise signal and an smooth average response to external stimuli.
However the usefulness of biological systems in electronic-like networks faces many challenges:
- In real-world applications, the big microscopy equipment will have to be scaled down.
- The time lapse between the stimuli and the change in read-out from the system must be reduced. Here the minimum response time is limited by the expression of a protein, which takes about 20 minutes.
- The nature of the read-out needs to be optimized: obtaining for instance an electronic rather than an optical signal, with bacteries emitting electrons directly.