Reference: Angew. Chem. Int. Ed. 2012, 51, 8744-8747
Title: DNA/polymeric micelle self-assembly mimicking chromatin compaction
Authors: Kaka Zhang, Ming Jiang, and Daoyong Chen
Review by: Anna Venancio-Marques
Title: DNA/polymeric micelle self-assembly mimicking chromatin compaction
Authors: Kaka Zhang, Ming Jiang, and Daoyong Chen
Review by: Anna Venancio-Marques
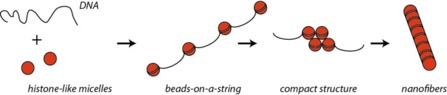
The authors show how to make tunable, monodisperse solenoidal nanofibers using DNA and micelles, with a sequence that is not unlike DNA interaction with histones to form chromatin fiber.
1. Getting to solenoidal nanofibers
- The authors use synthetic versions of histones to compact DNA and make the nanofibers. They mix DNA strands (here mainly 5427 bp), which are negatively charged due to the phosphate backbone, to positively charged micelles. These micelles are cationic spheres with an inert shell in PEG and a charged core (polymer called P4VP) that play the part of histone octamers.
- After compaction of the DNA around the micelles, the final nanofibers are made up of a fused core with DNA wrapped around the surface.
- The assembly is driven by electrostatic forces, to reach thermodynamically stable structures. This procedure gives rise to monodisperse objects when the starting material is also monodisperse.
- The authors use synthetic versions of histones to compact DNA and make the nanofibers. They mix DNA strands (here mainly 5427 bp), which are negatively charged due to the phosphate backbone, to positively charged micelles. These micelles are cationic spheres with an inert shell in PEG and a charged core (polymer called P4VP) that play the part of histone octamers.
- After compaction of the DNA around the micelles, the final nanofibers are made up of a fused core with DNA wrapped around the surface.
- The assembly is driven by electrostatic forces, to reach thermodynamically stable structures. This procedure gives rise to monodisperse objects when the starting material is also monodisperse.
2. Compaction steps
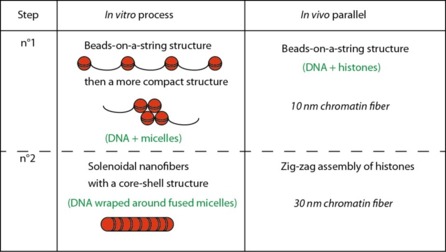
The first steps of the process of DNA compaction in cells and that achieved using micelles as histone-substitutes are very similar, sharing the same driving force: electrostatic interactions.
The first steps of the process of DNA compaction in cells and that achieved using micelles as histone-substitutes are very similar, sharing the same driving force: electrostatic interactions.
3. Achievements and perspectives
It is therefore clear that this paper provides a robust method for the synthesis of monodisperse, well-defined nanofibers with high throughput rates. The authors show it is also possible to monitor several parameters of the nanofiber by choosing the starting material carefully: starting with different DNA length gives rise to nanofibers of different final length, while varying the micelle size leads to nanofibers with different diameters.
The comparison of the in vitro step to the compaction around micelles is quite striking, as they share the sama driving force, electrostatic interactions, and lead to similar structure (beads on a string structure and then to more compact forms). The efficiency of DNA compaction achieved here is, however, much smaller than final DNA compaction in cells (10 000 for metaphase chromosomes, vrs a maximum of 8 for the nanofibers).
To develop even further the parallel with in vivo compaction introduced in the paper, it could also be interesting to study not only the sequence affinity of micelles, in the same way as DNA sequence influence the binding to histone octamer, but also the dynamics of this compaction process, for in cells the reversibility of DNA compaction is extremely important for transcription/replication purposes.
It is therefore clear that this paper provides a robust method for the synthesis of monodisperse, well-defined nanofibers with high throughput rates. The authors show it is also possible to monitor several parameters of the nanofiber by choosing the starting material carefully: starting with different DNA length gives rise to nanofibers of different final length, while varying the micelle size leads to nanofibers with different diameters.
The comparison of the in vitro step to the compaction around micelles is quite striking, as they share the sama driving force, electrostatic interactions, and lead to similar structure (beads on a string structure and then to more compact forms). The efficiency of DNA compaction achieved here is, however, much smaller than final DNA compaction in cells (10 000 for metaphase chromosomes, vrs a maximum of 8 for the nanofibers).
To develop even further the parallel with in vivo compaction introduced in the paper, it could also be interesting to study not only the sequence affinity of micelles, in the same way as DNA sequence influence the binding to histone octamer, but also the dynamics of this compaction process, for in cells the reversibility of DNA compaction is extremely important for transcription/replication purposes.
Further reading:
- to have easy-to-read material on DNA compaction in cells:
A. Annunziato, DNA Packaging: Nucleosomes and Chromatin, Nature 2008
- to see some of the pioneer work on DNA/histone-mimicking nanoparticles made by our group:
- to have easy-to-read material on DNA compaction in cells:
A. Annunziato, DNA Packaging: Nucleosomes and Chromatin, Nature 2008
- to see some of the pioneer work on DNA/histone-mimicking nanoparticles made by our group:
Compaction of Single-Chain DNA by Histone-Inspired Nanoparticles
A. A. Zinchenko, K. Yoshikawa, D. Baigl, Phys. Rev. Lett. 2005, 95, 228101 - doi : 10.1103/PhysRevLett.95.228101 ![]()
Single-chain compaction of long duplex DNA by cationic nanoparticles: modes of interaction and comparison with chromatin
A. A. Zinchenko, T. Sakaue, S. Araki, K. Yoshikawa, D. Baigl, J. Phys. Chem. 2007, 111, 3019-3031 - doi : 10.1021/jp067926z![]()