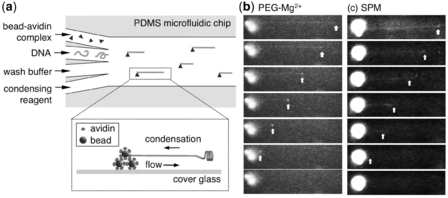
Figure 1: Single molecule compaction of DNA
Title : How environmental solution conditions determine the compaction velocity of single DNA molecules
Authors: Ken Hirano et al, Nucleic Acids Research, 2011, 1-6
Reviewed by: Naresh
Investigation:
Here the author studied the kinetics of compaction velocity of single DNA molecules using two non-protein condensation systems, Poly ethylene glycol (PEG) with Mg2+ and Spermine [1]. The viewers are requested to skim Prof. Yoshikawa’s website for elaborate explanations concerning DNA compaction induced by PEG [2].
Methodology:
The authors follow a simple methodology by creating a microfluidic channel (4 mm wide & 25 µm height) with branch channels (1 mm wide & 25 µm height) and injecting the bead avidin complex, DNA, wash buffer and condensing agents like PEG-Mg2+ and Spermine - Figure 1. (For more information: refer the article).
Authors: Ken Hirano et al, Nucleic Acids Research, 2011, 1-6
Reviewed by: Naresh
Investigation:
Here the author studied the kinetics of compaction velocity of single DNA molecules using two non-protein condensation systems, Poly ethylene glycol (PEG) with Mg2+ and Spermine [1]. The viewers are requested to skim Prof. Yoshikawa’s website for elaborate explanations concerning DNA compaction induced by PEG [2].
Methodology:
The authors follow a simple methodology by creating a microfluidic channel (4 mm wide & 25 µm height) with branch channels (1 mm wide & 25 µm height) and injecting the bead avidin complex, DNA, wash buffer and condensing agents like PEG-Mg2+ and Spermine - Figure 1. (For more information: refer the article).

Figure 2: Velocity of compaction of single DNA molecules
Interesting points highlighted in this article:
1. Explained the kinetics of structural transition of stretched DNA to compacted DNA at the single molecule level, though similar work has been carried out by Brewer et al using protamine as a condensing agent [2].
2. They elucidated the maximum averaged compaction velocity induced by PEG-Mg2+ and SPM as 60 micro m/s & 350 micro m/s (Figure 2).
Future perspective:
The protocol followed by the authors is robust and accomplishable. As a future perspective, it would be interesting to try transcription and helicase unwinding activity at single molecule level.
Brainstorming for the viewers:
S.G. Starodobtsev et al, already observed intra-chain segregation on single giant DNA molecule using Polyvinyl pyrrolidone (PVP) [4]. Does PVP would have a same effect on stretched DNA molecule? I am curious to explore this phenomenon.
References:
1. Ken Hirano et al., Nuceic acid research., 2011
2. http://www.chem.scphys.kyoto-u.ac.jp/
3. LR Brewer et al., Science., 1999, 286 (5437)
4. S.G. Starodoubtsev., Journal of Physical Chemistry., 1996, 100 (50)
1. Explained the kinetics of structural transition of stretched DNA to compacted DNA at the single molecule level, though similar work has been carried out by Brewer et al using protamine as a condensing agent [2].
2. They elucidated the maximum averaged compaction velocity induced by PEG-Mg2+ and SPM as 60 micro m/s & 350 micro m/s (Figure 2).
Future perspective:
The protocol followed by the authors is robust and accomplishable. As a future perspective, it would be interesting to try transcription and helicase unwinding activity at single molecule level.
Brainstorming for the viewers:
S.G. Starodobtsev et al, already observed intra-chain segregation on single giant DNA molecule using Polyvinyl pyrrolidone (PVP) [4]. Does PVP would have a same effect on stretched DNA molecule? I am curious to explore this phenomenon.
References:
1. Ken Hirano et al., Nuceic acid research., 2011
2. http://www.chem.scphys.kyoto-u.ac.jp/
3. LR Brewer et al., Science., 1999, 286 (5437)
4. S.G. Starodoubtsev., Journal of Physical Chemistry., 1996, 100 (50)